Evolution in Three Easy Steps: Carrying Out Directed Evolution
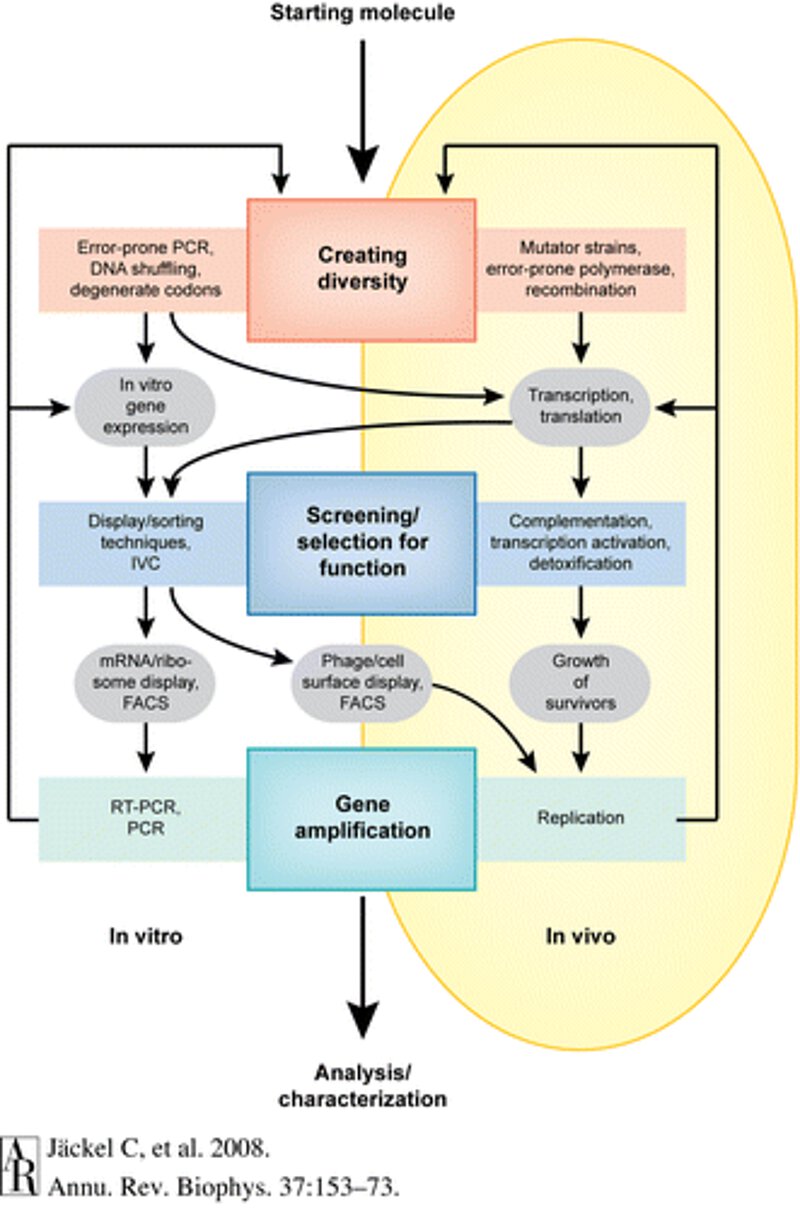 An overview of the steps in directed evolution.
An overview of the steps in directed evolution.
Directed evolution is emerging as a powerful new tool to design proteins with new and interesting properties that may not have evolved naturally. In the previous post in this series, we introduced the concept of directed evolution, and explained why it is useful. In this post, we discuss how directed evolution is carried out. There are three basic steps: 1) pick a starting protein, 2) create diversity, and 3) apply selective pressure.
1. Selecting a Starting Protein
As mentioned in the previous post, it is very difficult to design a new protein completely from scratch, because we don’t know enough about the relationship between the amino acid sequence and the final protein structure. Mutations far away from the active site can influence protein function, and could render the protein non-functional. That is why directed evolution involves starting with a functional protein. The protein in question must be adaptable, otherwise it will not be able to evolve at all, not even through natural selection! A good way to tell whether a starting protein is a good candidate for directed evolution is to check its natural functional diversity: if a protein has a wide variety of functions across its family, chances are that it will successfully “evolve” in the laboratory.1
2. Creating Diversity
The sequence of the starting protein needs to be mutated to form a library of new sequences. This library can be generated in a number of ways. The sequence can be randomly mutated in vitro through error-prone PCR or DNA shuffling, which introduces mutation through recombination. GenScript provides services to generate a variety of mutant libraries. In vivo mutations can be obtained in a variety of ways. For example, when activated by an antigen, B cells can quickly introduce mutations in their antibody genes using a process known as somatic hypermutation. Foreign genes introduced into B cells can also be mutated by somatic hypermutation.2 Finally, several vendors at Scientist can help generate libraries using computational mutagenesis.
3. Selective Pressure
After generating a library of new protein sequences, it’s time to select which ones will work the best. One way of doing this is to examine each sequence individually by screening the library with high-throughput functional assays, and select the highly-functioning mutants. Another way is to test all the sequences at the same time is by introducing selective pressure, thereby eliminating all non- or poorly-functioning mutants.
Lather, rinse, repeat
Once the “fittest” mutant is found, it is used as the starting protein for the next round of directed evolution, and the process is repeated until all the desired properties are introduced into the protein.
In the final post of the series, we talk about applications of directed evolution.
References- Romero, PA and Arnold, FH. Exploring protein fitness landscapes by directed evolution. Nature Reviews Molecular Cell Biology 10 (12): 866-76. December 2009.
- Brindle NPJ, et al. Directed Evolution of an Angiopoietin-2 Ligand Trap by Somatic Hypermutation and Cell Surface Display. J Biol Chem, 288(46):33205-12