Bringing Clarity and Efficiency to Early Drug Screening: How Chemical Protein Stability Assays (CPSA) Help Drive Decisions at the Hit/Lead Discovery Stage
This blog post was written by Medicines Discovery Catapult, an independent drug discovery innovation centre reshaping drug discovery for patient benefit. Their services are available on Scientist.com.
Early-stage drug discovery hinges on understanding whether potential compounds truly engage with their intended protein targets. Traditional approaches to confirming these interactions often involve complex workflows, purified proteins and label-dependent assays, each introducing potential bottlenecks and variability.
The Chemical Protein Stability Assay (CPSA), developed by Medicines Discovery Catapult offers a smarter alternative. This proprietary, lysate-based, mix-and-read solution is designed to bring clarity, efficiency and physiologic relevance to early-stage drug screening. By delivering label-free, high-throughput target engagement data, CPSA helps researchers make faster, more confident decisions about which compounds to prioritize.
Simplifying Protein – Ligand Binding Assessment
At its core, CPSA enables scientists to rapidly assess cellular protein – ligand or protein – drug binding through a scalable, accessible workflow. It uses a cell lysate – based chemical denaturation technology to detect protein stabilization induced by ligand binding, confirming target engagement in a single, straightforward assay.
Unlike conventional techniques that require multiple purification or transfer steps, CPSA operates entirely within one plate in a mix-and-read format. This simplicity eliminates sources of assay variability and reduces overall costs. Whether a team is evaluating a handful of lead compounds or conducting large-scale 1536-well library screens, CPSA adapts seamlessly to the throughput needed.
Moreover, because CPSA does not depend on target enzymatic activity, it can identify ligands that bind outside the active site, unlocking new chemical space and supporting the discovery of novel molecular series.
How Does CPSA Work, and Why Does it Deliver Better Data?
CPSA is based on measuring the stabilization effect a ligand has on a target protein when exposed to chemical denaturants. Proteins naturally unfold under denaturing conditions — but when bound to a ligand, they resist unfolding, indicating true target engagement.
This approach provides several critical advantages:
- Label-free analysis: no need to modify proteins or ligands, ensuring that the molecule’s true pharmacology remains intact.
- Single-well simplicity: eliminates plate-to-plate transfers, reducing variability.
- Scalability: easily configurable for both dose-response and single-point screening.
- Physiological relevance: cell lysate provides a native protein environment, avoiding challenges of large-scale protein purification.
“CPSA was designed to bring clarity and efficiency to early-stage screening. By combining a straightforward workflow with label-free, physiologically relevant data, we’re helping researchers make faster, more confident decisions about their lead compounds.” - John Vincent, Lead Scientist, Assay Development
Key Advantages of Chemical Protein Stability Assay
- Plate reader – based assay accessible to most research labs.
- Simple, scalable workflow (compatible with 384 – 1536 well plates).
- No high-temperature incubations required.
- No lysis or material transfer steps, improving reproducibility.
- Uses native protein lysate, maintaining biological context.
- Cost-effective, label-free approach.
How Does CPSA Compare to Standard Protein Stability Assays?
Traditional protein stability assays typically involve high-temperature incubations, lysis steps and multiple plate transfers. These processes are not only time-consuming but can also lead to inconsistencies in data quality. The requirement for purified protein further limits throughput and introduces potential artefacts from non-native environments.
Standard Target Engagement Assay Workflow
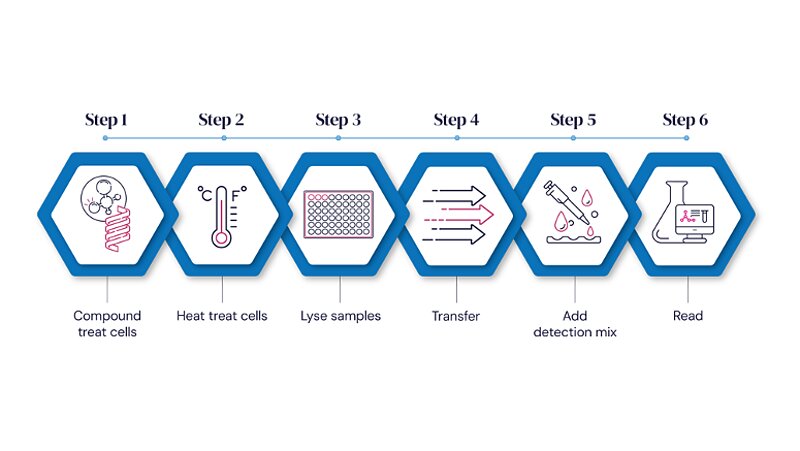
CPSA simplifies this entire process. By employing cell lysates instead of purified proteins, CPSA removes the need for large-scale protein production and avoids heat-induced denaturation. Its single-plate, mix-and-read workflow minimizes handling steps and experimental variability, making it ideally suited for high-throughput screening (HTS) and rapid lead profiling.
CSPA Workflow
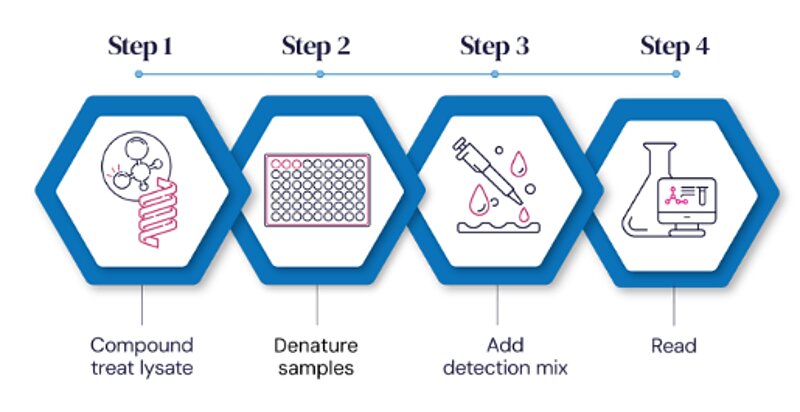
In practice, CPSA allows scientists to generate robust, reproducible target engagement data with fewer resources and in significantly less time than traditional methods.
“CPSA is an ingenious extension of the target engagement toolbox. It is easier to upscale and control in high-throughput screening applications than currently available methods. With exemplary support from MDC, we have smoothly incorporated it into our Drug Discovery pipeline.” - Selvita, Maciej Olszewski, PhD, DSc, Associate Director, In vitro Pharmacology and Translational Research
Driving Discovery Forward
CPSA can be applied throughout the drug discovery continuum - from hit identification to lead optimization, across diverse protein targets and cellular matrices. Its flexibility and physiological relevance make it a valuable component of the modern drug discovery workflow.
By combining scientific depth with operational simplicity, CPSA represents a major step forward in how researchers confirm and interpret target engagement. It enables more meaningful comparisons between compounds, improves decision-making speed and accelerates progress toward identifying true drug candidates.
Ready to simplify your screening workflow and unlock more meaningful engagement data?
Click here to learn more about how CPSA can support your next discovery project.